
The labels “Create” and “Remove” are now used consistently throughout MacVector for all buttons that add/create or delete/remove features. The Feature tab context menu now lists all /db_xref and /protein_id qualifiers – selecting one will open the appropriate database source in the default web browser.
The Feature tab has a new context sensitive (right-click) menu item that lets you join two or more selected features of the same type.

MACVECTOR PROTEIN TOOLBOX CODE
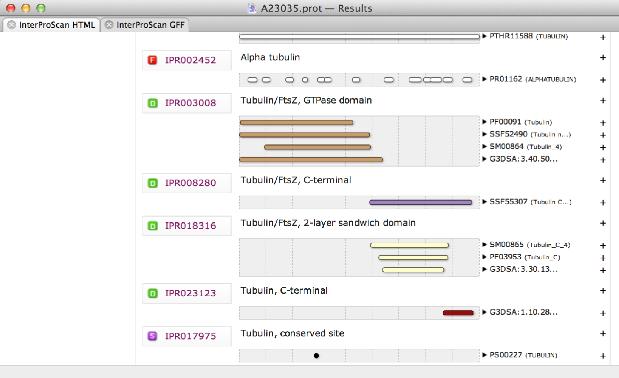
There is a new Genetic Code reference window accessed from the Window | Genetic Code Key menu item that displays the codons of the currently selected genetic code for quick reference. Align to Folder – now scans hierarchical sub-folders as well as the selected folder. This represents matching residues in an alignment with dots rather than the residue itself. There is a new “Show Dots” button that can optionally be added to the MSA editor toolbar (right-click on the toolbar and choose Customize Toolbar… to add the button).
MACVECTOR PROTEIN TOOLBOX WINDOWS
These changes let you easily paste primers from external applications or from other MacVector windows into a subsequence file to maintain a database of useful primers. The Subsequence editor has been reworked as a dropdown dialog sheet and you can now copy/paste into the Sequence edit box.
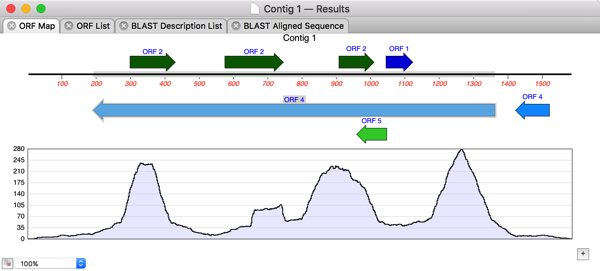
NA/AA Subsequence – the graphical Map result window is now linked back to the parent sequence so that selections in the result window are propagated to the parent editor. In addition, the pI and molecular weight calculations have been revised for increased accuracy. Protein Analysis Toolbox – the AA Composition, pI and MW protocol now also displays the predicted extinction coefficients and aliphatic index. The selected enzyme data file is now displayed in a history control so you can quickly swap between commonly used files. Restriction/Proteolytic Enzyme – you can now specify the minimum number of cuts as well as the maximum number of cuts when filtering (or setting up) the results. The Ligation dialog now treats sequences copied from external sources or the sequence editor as blunt-ended fragments. a feature) and Edit | Copy will copy a blunt ended copy of the underlying sequence to the clipboard. You can now select any graphical object (e.g. Microsoft Word or Text Edit (OS X 10.5 and above only). TextEdit or Microsoft Word) that can accept drag and drop text.You can also drag selected text out of a result window and drop it into other acceptor windows e.g. You can now drag a selection from one sequence window and drop it into another single sequence window or into an external application (e.g. All three have also had extensive work to streamline workflows, add keyboard navigation and fix various display glitches and crash bugs.ĭialogs to locate auxiliary data files (Restriction Enzymes, Matrices, Codon Bias tables etc) now all default to the appropriate location in the MacVector distribution folder.Īlign to Folder, Restriction Enzyme, Proteolytic Enzyme, Nucleic Acid Subsequence, Protein Subsequence, Translation and Reverse Translation are now all window modal dialog sheets. Multiple levels of Undo have been added to the Trace, Multiple Sequence Alignment and Contig/Align to Reference editors. Many of the sheets have a convenient new history combo box that keeps track of the various data files or target folders used in the dialog to allow rapid switching between commonly used files. Many of the analysis functions now use drop down window-modal sheets rather than application modal dialogs, allowing you to continue to work with other windows to prepare, view or copy data required for the analysis. You might also want to read our blog article discussing the new release. A more complete list of changes is present in the MacVector 11.1.1 Release Notes. MacVector 11.1 has hundreds of small enhancements and bug fixes designed to increase stability and improve workflows and the user experience. MacVector is widely regarded as the most intuitive, easy to use program available for sequence analysis.Sequence Analysis Tools for Molecular Biologists


 0 kommentar(er)
0 kommentar(er)
